Evolution is a lot messier than we thought
Cells evolved haphazardly, not in one overall arc
At least a thousand million years ago, two unicellular organisms collided and become one. Like mismatched Russian nesting dolls, one ancestor of modern bacteria became a compartment inside an ancestral eukaryote, one of the complex cells that eventually went on make up all animals, plants, and fungi. The ancestral bacteria evolved into mitochondria, which nowadays generate energy for all eukaryotic cells, using genes inherited from their former bacterial lifestyle. Modern mitochondria are relics of this ancient endosymbiosis event – when two lineages teamed up into one organism – and today they still hold a record of how this relationship developed over time.
Scientists, who are eukaryotes themselves, have long taken a eukaryotic-centric view of eukaryotic evolution. That cells developed internal compartments with specialized functions was an unexpected twist and a major advance for life on Earth. Mitochondria are thought to have either triggered or coincided with the earliest stages this process. They also have smaller genome sizes than larger eukaryotes, making them convenient markers for studying the evolutionary distance between their hosts. Until recently, all known modern mitochondria showed the same record: a straightforward path from wild bacteria to domesticated endosymbiont.
Life without known relatives
Recently scientists discovered a new species of single-celled organism, without known relatives, that has challenged the view that mitochondria evolved along one path. Instead, haphazardly but successfully, mitochondria have been evolving independently, along different lineages, each solving the problems of being an endosymbiont in their own way.
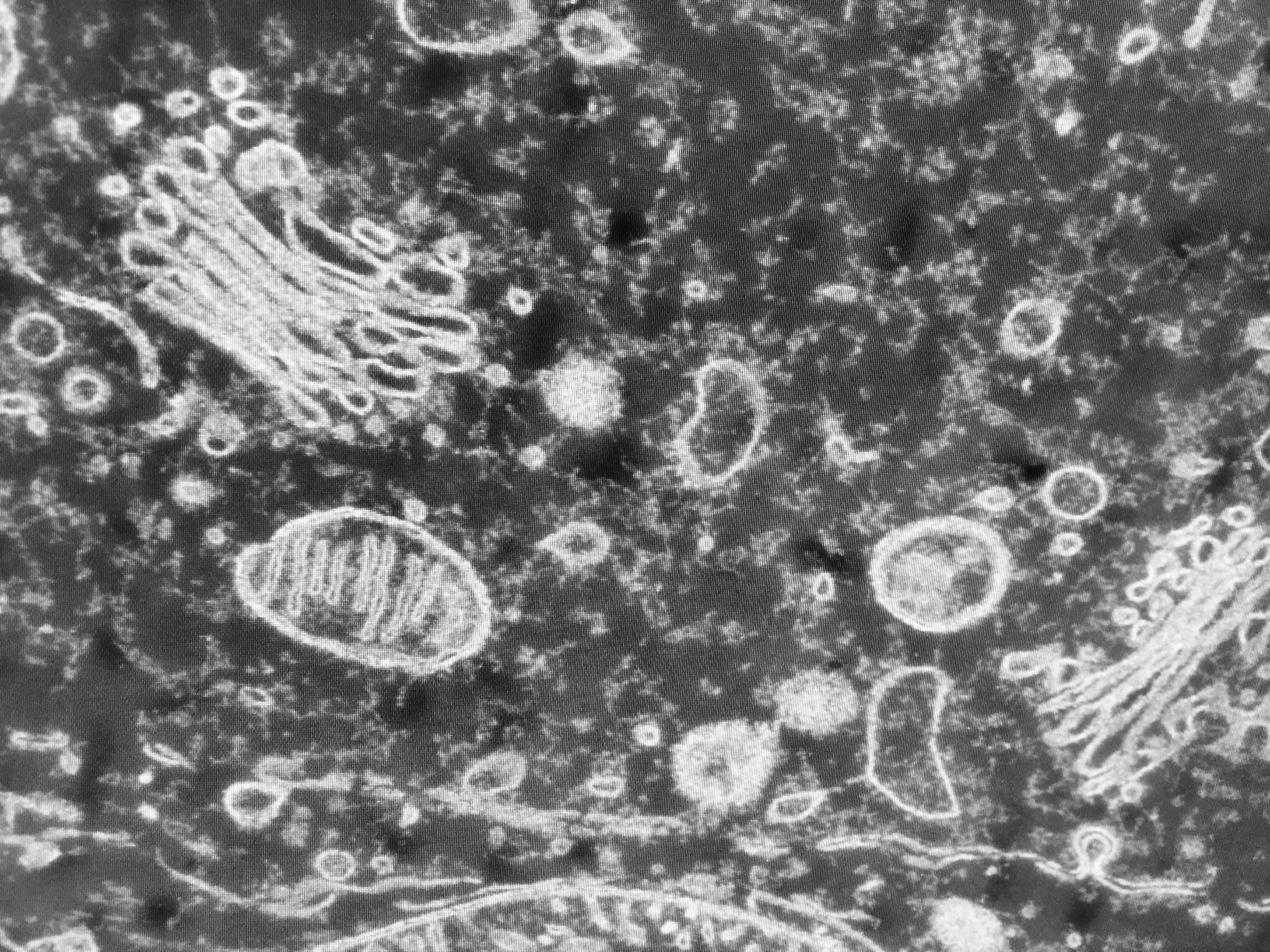
Mitochondria in a cell
Just as scientists have tracked how bacteria adapt to a specific environment – the human gut, soil, the laboratory – by examining changes to their DNA, evolutionary biologists have a long-standing interest in investigating mitochondrial DNA. But the mitochondrial genome has changed in ways that are unique to its lifestyle as an endosymbiont: it has relocated huge chunks of its genome to its host cell's nucleus, leaving it with only a paltry number of genes inside its mitochondrial membranes.
What drove this massive restructuring? Scientists believe that the health of the host cell, which relies on energy produced by mitochondria, was the ultimate cause. They have hypothesized that relocating mitochondrial genes to the nucleus removed the burden on mitochondria of synthesizing extraneous proteins, for instance, or it may have relieved spatial constraints in the cell. Whatever the causes along the way, the results are stark: the modern relatives of mitochondria have thousands of genes, while no known modern mitochondria have held on to even one hundred.
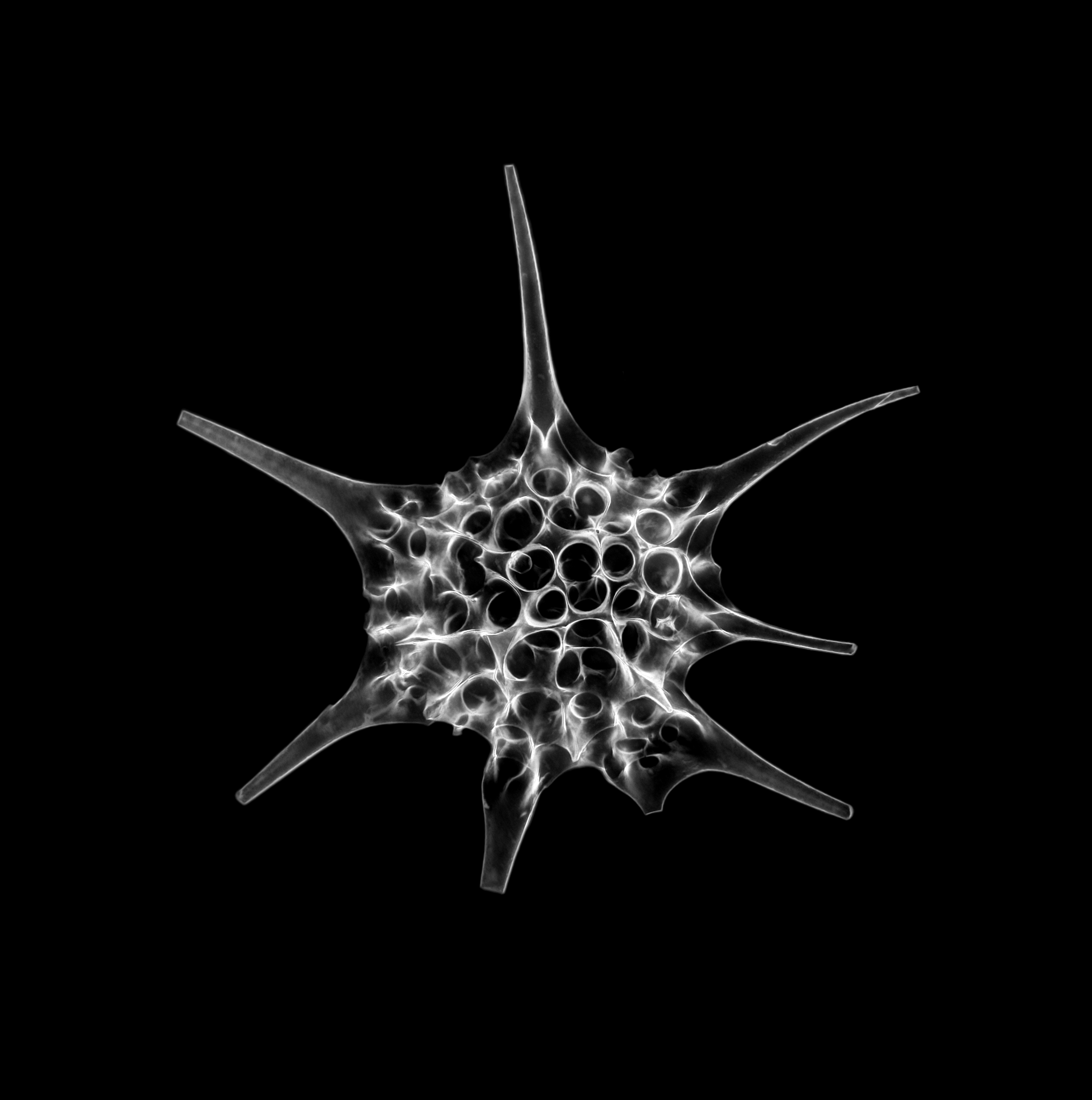
A protist
This massive downsizing is most extreme in animal mitochondria. Nearly all mitochondria in the animal lineage share the same tiny number of genes (only 13 protein coding genes). Scientists have used these few genes as a powerful tool, examining small rearrangements and minor mutations to measure the evolutionary relationships between animals. But while useful, these differences are mostly cosmetic. To learn how mitochondria transitioned from wild bacteria to domesticated endosymbionts, biologists are looking outside of the animal lineage, where the process of genome reduction is still ongoing. A newly discovered species suggests genome relocation is far more dynamic and messy than scientists previously thought.
Ancoracysta twista is a newly described, single-celled organism that has the lonely distinction of having no known evolutionary relatives. For a single-celled organism – in a category called protists – A. twista leads an exciting life: found on a brain coral in a tropical aquarium, it has a flagella for swimming and is thought to stab and immobilize prey using a tiny, extruding hooked spear. This and other novel morphological features, recently described in Current Biology by Jan Janouškovec and a team at the University of British Columbia, reflect a unique evolutionary trajectory: a creature branching off the tree of complex cellular life, apparently on its own.
But A. twista’s most interesting feature may be its mitochondria, which contains one of the largest mitochondrial genomes seen to date with more in common with modern bacterial relatives. The other organisms with large mitochondrial genomes are also protists; strikingly, they are not related to A. twistaand their mitochondria contain non-overlapping sets of genes. This difference suggests that mitochondria did not relocate their genome in one jump, but multiple times, independently, in different lineages of life.
Dumpster diving through genomic trash
Searching for further evidence of this pattern, Janouškovec's team turned to the trash can of genome assembly. When genomes are sequenced, the data output is billions of tiny fragments of DNA. A computer algorithm then stitches these together based on their overlapping ends, in a process called genome assembly. Some DNA fragments are left out, and are consequently thought to originate from a different source, such as an accidental contaminant or the mitochondrial genome. By mining these discarded sequences, Janouškovec reconstructed the mitochondrial genomes of even more unrelated protists. They found that they, too, had large, imperfectly overlapping genomes.
Altogether, the data shows these major rearrangements actually say very little about how closely related two species on the evolutionary tree. Instead, each lineage has a unique story about how their particular mitochondria evolved.
Not one path but many
The process of relocating genes from the mitochondria to the host nucleus is an extremely complex problem, taking many steps and a lot of evolutionary time. Although mitochondria have the DNA instructions to make only a small number of proteins "in house," ultimately they contain over a thousand different proteins needed for various functions. Therefore, genes that were historically encoded by the mitochondria must be expressed by the host cellular machinery and then re-imported to the mitochondria, via a dizzying assortment of sorting and uptake machinery. Amazingly, the clues in mitochondrial DNA show us that cellular life has overcome these challenges, entrusting important genes to a distant nucleus, not once but possibly many, many times.

Another protist
The large genomes of A. twista, and other protists, each provide a different snapshot of the possible paths that animal mitochondria passed through, as they downsized to their modern reduced form. Where these paths converge suggest the requirements for mitochondrial domestication; where they diverge highlights where endosymbiosis tolerates messiness. Each is important for understanding how two single-celled organisms slowly developed, from that first collision to today, into their current endosymbiotic relationship.
As genome sequencing becomes less and less expensive, our picture of the biological present, and how it was influenced by the biological past, can and should expand to include new organisms, like the strange and tiny A. twista. Protists are one of the most abundant and understudied forms of life on earth, and many scientists are calling for greater sequencing efforts to capture their diversity. Indeed, the insight that major changes in mitochondrial DNA are not simple markers of evolutionary relatedness, and that evolution within the cell is as messy as evolution outside it, came from probing the far reaches of the evolutionary tree.




I enjoyed this piece! A few questions:
1. Is endosymbiosis the name of the theory that proposes this merging? And/or does it mean "symbiosis from within the host"?
2. what are the mechanisms for the dramatic reduction of mitochondrial genomes? How does it transfer DNA and manage the gain/loss of proteins between the two organelles?
3. With concerns in off-target genetic editing and the attempt to build an intelligently designed yeast genome from scratch, understanding why and how cells organized (or failed to control) all these gene transfers could have some potential benefit to these applications...